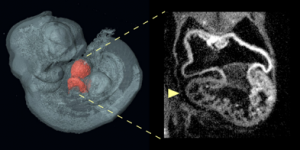
An optical projection tomography of an E10.5 Nipbl+/- embryo shows on the left a three-dimensional reconstruction of embryo with heart highlighted in red; on the right is optical section through the heart showing abnormally small right ventricle in the Nipbl+/- mouse.
Credit: Benedikt Hallgrimsson/University of Calgary
New research published by Professors Anne L. Calof, Anatomy and Neurobiology and Developmental and Cell Biology; Arthur D. Lander, Developmental and Cell Biology; and their colleagues, report that the role of genes in congenital heart defects (CHDs) is more complex than previously realized, and that overall risk of the disease is determined by a combination of gene effects both inside and outside of the heart itself.
Formation of a normal heart depends on the interactions of multiple types of cells that collaborate in precise times and places throughout development to build the heart’s intricate structures. To figure out how these interactions can go awry, the Calof-Lander team studied atrial septal defects, a common type of heart defect, in a mouse model of the developmental disorder Cornelia de Lange Syndrome (CdLS).
Cornelia de Lange Syndrome can be caused by mutations that inactivate a single copy of Nipbl, a gene that directs the expression of many hundreds of other genes in tissues throughout the body. Using genetically modified mouse models, the Calof-Lander team used a novel technology to selectively introduce or remove Nipbl mutations in different tissues during embryonic development. The group found that Nipbl deficiency in no single tissue could single-handedly account for the development of atrial septal defects. Rather, the development of heart defects was determined by interactions between heart-forming tissues and the rest of the body.
“When a single gene change causes a birth defect, we often assume that it’s because one thing goes wrong in one cell type. The big difference in our studies may have to do with the fact that Nipbl controls a large number of other genes,” said Lander. “Given that most human CHDs are now thought to be caused by gene variants acting in combination, what we learned from Nipbl-deficient mice may actually be more typical of the way most CHDs arise.”
Other researchers who contributed to the study were: Rosaysela Santos, Shimako Kawauchi, Russell Jacobs, Martha Lopez-Burks, Hojae Choi, Jamie Wikenheiser, Benedikt Hallgrimsson, Heather Jamniczky, and Scott Fraser. The complete study results can be found online in PLOS Biology.
